A socratic dialogue over the utility of DNA language models (Part 2 of 2)
4.9k words, 23 minute reading time
Part 1 is focused on variant pathogenicity prediction using these models.
Part 2 is focused on genome generation using these models.
I am aware that DNA language models are useful for things other than those two (like protein fitness), but variant prediction and genome generation are the two bits that I find most interesting.
Twitter link for this article.
Introduction
(The Introduction is a repeat from Part 1. Skip it if you’ve already read it!)
I think I, alongside many other people in this field, live in this seemingly parallel universe where we don’t really understand why anyone is working on DNA language models. I say ‘parallel’, because there is obviously a world in which some very smart people are very much bullish about them: specifically the Arc Institute. Who, just yesterday, released a paper that many people are quite excited about: Evo 2, a successor to the original Evo model. From the news article:
Arc Institute researchers have developed a machine learning model called Evo 2 that is trained on the DNA of over 100,000 species across the entire tree of life. Its deep understanding of biological code means that Evo 2 can identify patterns in gene sequences across disparate organisms that experimental researchers would need years to uncover. The model can accurately identify disease-causing mutations in human genes and is capable of designing new genomes that are as long as the genomes of simple bacteria….Building on its predecessor Evo 1, which was trained entirely on single-cell genomes, Evo 2 is the largest artificial intelligence model in biology to date, trained on over 9.3 trillion nucleotides—the building blocks that make up DNA or RNA—from over 128,000 whole genomes as well as metagenomic data. In addition to an expanded collection of bacterial, archaeal, and phage genomes, Evo 2 includes information from humans, plants, and other single-celled and multi-cellular species in the eukaryotic domain of life.
Which on face value seems cool and obviously impressive. Right? You read this and instinctively go, ‘wow, neat!’. But if you then try to ask ‘why would anyone want this?’, there aren’t many resources to go to (though I did enjoy Asimov’s recent piece about it). At least outside of the paper, which is a bit hard to immediately understand. For protein folding, the use case of something like Alphafold2 was obvious: if you know the shape of a protein, you can design things that bind to them. And even use the model to generate binders! If you have a model that has a really good latent understanding of DNA sequences, you can…do what exactly? Maybe for some people, the answer is obvious. But it’s not for me, someone who isn’t a genomics person. So this post will be discussing that.
I learn best via Socratic dialogue, which is just argumentative dialogue between individuals based on asking and answering question. This post will have that same setup, but for DNA language models. Notably, this is not specifically a post about Evo 2 (though I will often refer to it). It is more about what utility such a model would even have. So, no data or architectural details here, outside of where necessary to discuss use cases.
The dialogue
Okay, so, previously we talked about how DNA language models are useful for variant pathogenicity. It’s an open question how useful it’ll end up being, but I can see the argument there. But there’s a second part to the Evo 2 paper: the utility of it for genome generation. I don’t understand this at all. Why would you want to generate genomes?
Remember how, when we discussed variant pathogenicity, we drew this line connecting conservation scores and Evo 2 log likelihoods? It may be helpful to do something similar here for genome generation.
Do people ever actually do genome generation?
Somewhat. To start off with, let’s focus on partial genome generation. We’ll come back to full genome generation later on.
The most basic case of genome generation can occur when people need to fill in gaps in sequencing data. Next-Generation-Sequencing (NGS) data involves chopping up DNA into small fragments, reading those fragments, and then trying to stitch them back together. Sometimes, there are regions that just don't get sequenced well—maybe they're too repetitive, or too GC-rich, or [something else]. That’s why some parts of the human genome remained technically incomplete until just a few years ago.
A model like Evo 2 could potentially generate biologically plausible sequence for those gaps, based on the surrounding context. Definitely not something that’d be considered ‘completing a genome’, but, just as is the case with variant pathogenicity, it helps researchers start from somewhere as opposed to nowhere.
That sounds like a terrible use-case. Imputing the rest of a genome given fragments of it feel like the sort of thing where nearly perfect accuracy really matters, and I’m not sure I’d trust an autoregressive model like Evo 2 to get there.
You’re right! And that’s why the authors don’t even bother to test Evo 2 on that, it’s not a particularly compelling problem, especially since there are plenty of other methods, like long reads with nanopore sequencing, to side-step the whole issue. But it’s a good mental stepping block!
Now, let’s go slightly further. A more complex place people do genome generation is by trying to get the cell to produce a molecule of interest, say, insulin. They wrap the INS gene into a plasmid, force it into the yeast cell, and it starts to produce insulin.
Okay, but that also doesn't really count. That's just inserting a gene we already know into a cell, which doesn’t even integrate with the yeast’s genome. There's no generation happening, just copying and pasting. And before you say, ‘you need to do codon optimization also’, that also doesn’t count! That’s just a lookup table,"CAG becomes CAA" and so on. It's still hardly "genome generation" in any meaningful sense.
You’re wrong! In practice, the way yeast cells are told to produce insulin does involve a fair bit of engineering beyond pure codon optimization.
Remember, in all cells, insulin isn’t produced as insulin right away. It starts as preproinsulin, which then gets processed into proinsulin, and finally into active insulin. In human pancreatic beta cells, preproinsulin has a built-in signal peptide at the very beginning of the protein sequence to cause this chain of events. This signal peptide tells the cell to send the protein into the endoplasmic reticulum (ER), where it gets processed into proinsulin and eventually into mature insulin. Once preproinsulin enters the ER, the signal peptide gets cleaved off, and the remaining protein continues along the processing pathway.
But yeast doesn’t have the same protein processing machinery as humans do. So if you want yeast to secrete insulin, you can’t just rely on the human version of the signal peptide. Instead, you need to swap it out for a yeast-specific signal peptide—one that yeast actually recognizes and responds to. And, unlike codon optimization, there wasn’t a lookup table for this sort of work, since yeast doesn’t naturally produce insulin! It took years of work to nail down the most optimal signal peptide to use.
But even then, that’s still simple! It’s just a peptide you need to modify on a pre-existing INS gene, which, while annoying to tune, still has a reasonably small search space.
Fair, so let’s now consider a genome generation task that is actually complicated.
Consider antibodies. Antibodies are really, really hard to make. This is partially because you typically need mammalian cells to produce them, which are notoriously easy to kill, hard to genetically modify, and grow slowly. Why can’t we use yeast, like we do for insulin? I’ve discussed this before in an antibody article, quoting from here:
Yeast is challenging to kill, replicates easily, grows fast, and is amenable to genetic manipulation. So why don’t we use them [to create antibodies]? There’s a really wonderful review paper that discusses all this. Antibodies require specific post-translational modifications, particularly glycosylation, at a specific residue (residue 297 of each heavy chain), to function ideally in the human body. While yeast cells do have their own glycosylation machinery, it differs substantially from that of mammalian cells. Yeast tends to add high-mannose type glycans (as in, sugar molecules that contain a lot of mannose) to its produced proteins, which are not typically found on human proteins and can potentially make the antibody more immunogenic, which is obviously undesirable. Looking beyond yeast has similar issues; many types of bacteria lack any glycosylation system at all.
So are we doomed? Well, no. You could do something called ‘glycosylation engineering’ on the yeast cell, which would hopefully push our yeast cell into producing more human-like sugars.
The obvious question is…how exactly do you do this?
Simple: knock out yeast-specific glycosylation enzymes that add high-mannose sugars, introduce human glycosylation enzymes that add the correct sugars in the correct locations, modify regulatory elements so the new pathway is expressed at the right levels, and, of course, ensure that all the above changes don’t affect the capacity for the yeast cell to continue surviving, reproducing, and secreting the antibody.
That doesn’t seem simple at all.
It’s sarcasm, which understandably doesn’t come across well through text. But yes, it’s hard. It shouldn’t be surprising that glycosylation engineering as a field is very much in its infancy, precisely because the genetic engineering you need to perform involves replacements much of a yeast cells genome. And there is no simple way to do that! There are tons of genome generation tasks like this that people have to do almost entirely manually or are just left completely unexplored because it’s so hard to explore the search space.
Okay, that’s a complex-enough task that I’d be comfortable identifying it as ‘genome generation’. But what you’ve just described is a conditional form of genome generation, where you’re trying to push a yeast’s genome into a specific direction, that direction being ‘use human-like sugars’. Returning back to Evo 2, the model seems like it’s only capable of conditional generation in terms of surrounding nucleotides, but it cannot be steered towards a specific function. All it can do is make ‘natural’ looking DNA. Am I wrong?
You’re right, but as with all generation processes, guiding them isn’t too hard if you have access to a good-enough oracle to sift through the generations. And in fact, the Evo 2 authors did exactly that.
Some quick background: some parts of a genome are open chromatin, meaning they’re accessible to transcription factors, polymerases, and other regulatory proteins. These are typically active promoters, enhancers, and regulatory regions that influence how genes are expressed. Other parts are closed chromatin, meaning they’re tightly packed and essentially invisible to the transcription machinery. These tend to be silenced genes or non-functional regions. Why isn’t everything open? Because not all cell types need access to all genes; a liver cell does not need the same proteomic machinery as a cardiac cell.
This pattern of open/closed are also often referred to as the ‘chromatin accessibility profile’ of a genome. While one would be correct in referring to this as an epigenetic feature of a genome — as in, a chemical marker on top of the nucleotides, and not the nucleotides themselves — it’s also true that the nucleotide sequence can predispose certain regions to accessibility or inaccessibility. Thus, if you have access to the sequence of the genome, you have access to a fuzzy view into its chromatic accessibility profile.
Returning back to the subject at hand, Evo 2 was used to redesign a genome’s chromatin accessibility profile. The following strategy was employed:
Start with a small prompt sequence (in this case, a few thousand base pairs from a mouse genome).
Generate multiple sequence continuations (128 bp chunks at a time) using Evo 2.
Evaluate each generated chunk using external chromatin accessibility prediction models (Enformer and Borzoi).
Keep only the best-scoring chunks—i.e., the ones whose predicted chromatin accessibility matches the desired pattern. Regenerating the nucleotide continuations from 2 over and over again (up to 60 times) until these models gave the green light led to increased performance.
Repeat the process until a full sequence (~20 kb) is generated.
This is a complex task! It isn’t perfectly nailed down exactly how to modify chromatin accessibility, but we do know that certain motifs and sequence patterns contribute to it. Transcription factors (TFs) play some role here; if a sequence has a strong TF binding site, it’s more likely to have an open chromatin profile. Conversely, certain DNA structures — like tightly packed nucleosomes — can make a region inaccessible, making it have a closed chromatin profile. But there’s no lookup table for this sort of work.
What was the actual problem that Evo 2 was tasked with?
The specific task assigned to Evo 2 was to generate DNA sequences that encode predefined chromatin accessibility patterns, including “EVO2” (. ...---- ..---) and “ARC” (.- .-.-.-.), along with a few related designs. In this encoding scheme, open chromatin represents dots and dashes, while closed chromatin represents spaces. Each character corresponds to 384 base pairs, equivalent to three sequential Evo 2 generations.
If you’re confused by the referring to this as ‘morse code’, so am I. Practically speaking, something like EVO2 translates to open-closed-open-closed, and ARC translates to open-closed-open. Which feels somewhat clumsy, but it’s a minor point.
And how did they assess performance?
This is where at least a few people have been somewhat bewildered. The authors rely entirely on the chromatin accessibility method models to evaluate the success of their designed sequences—specifically, whether the predicted accessibility pattern aligns with the intended Morse code signal. In other words, they generated the sequences, fed them back into Enformer and Borzoi, and checked if the models agreed that the correct regions were open or closed.
So, no experimental validation? Evo 2 is optimizing its outputs based on Enformer and Borzoi, and then its success is measured by Enformer and Borzoi? Isn’t that a clear case of adversarial optimization?
Well, maybe. The authors do touch on this a little. Here’s a plot:
So, figures E and F come from nucleotides generated via Evo 2. Figure E says that that Borzoi and Enformer are confident and accurate about their prediction of open/closed chromatin regions — at least, if you trust the models predictions. And Figure F says that the distribution of generated dinucleotides (so, pairs of nucleotides, of which there are 16) match up with the pattern of typical mouse genomes.
To study this the possibility of adversarial optimization, the authors compare the Evo 2 generated nucleotides to nucleotides that were generated through an entirely random process (and still filtered via Enformer and Borzoi, and sampled at high rates). Here is that plot:
Figures B and C are analogues to figures E and F. Here are the two fundamental claims that the authors make from these plots:
If Evo 2 produced adversarial sequences, you’d expect strong deviation between Enformer and Borzoi. You do see this deviation with uniform generation, but not with Evo 2.
If Evo 2 produced adversarial sequences, you’d see deviation from the naturalness of the produced nucleotide patterns to the reference mouse genome. You do see this deviation with uniform generation (at least for some patterns), but not with Evo 2.
Thus, Evo 2 is (probably) not falling prey to adversarial generation.
I think, fairly, this wouldn’t be considered a satisfying enough explanation to most. But I do think there is an inkling of some assurance here, especially with 1. After all, RFDiffusion also relies on this same sort of ‘orthogonal selection’ by assessing generated proteins partially on the structural agreement between RFDiffusion and Alphafold2. And that seems to work well!
So why wouldn’t it work well for the nucleotide parallel?
Who knows? I’m not deep enough in the field to say whether Enformer and Borzoi are independent enough to serve as a valid cross-check. I also do not know whether ‘unnatural-looking’ dinucleotides are evidence of…anything really. I agree with basically everyone else that experimental validation will be necessary to trust these results further.
I think a particularly worrying bit is Figure A in that second figure you attached. Uniform seems to work…surprisingly well. It’s certainly worse than Evo 2, but not that much worse. So, all the fuss you made about the complexity of glycosylation engineering doesn’t transfer to this genome generation task. Empirically, remodeling chromatin accessibility doesn’t seem to be that hard!
Yeah…I don’t know. I think the reliance on trained oracles to use for filtering Evo 2 outputs means that, for now, it is a bit limited in the complexity of what it can do. I do wish the authors tried it out on a few, super low-shot genome engineering tasks, where uniform nucleotide generation completely fails. Something like generating a minimal enhancer that can activate a reporter gene in a specific cell type or designing a simple promoter that works across multiple species. I have no idea what that would look like…maybe fine-tuning Evo 2 on a few cases of enhancers + promoters, and seeing how well it can make new versions?
I do think there will be a slew of follow-up papers that try to attack all the potential use cases here. Maybe something even equal to the complexity of glycosylation engineering! We’ll see, I can very much understand wanting to not make a 40 page paper even longer.
Okay, moving on. Thus far, we’ve only discussed partial genome generation, but there is also this take I keep seeing on Twitter about how DNA language models are useful for generating whole genomes outright. I don’t really grasp this at like…an object level. What does it even mean to generate an entirely new genome?
Well, it’s not an entirely new concept. People have made synthetic genomes for a while now, the most well-known and oldest one being the Mycoplasma minimal genome project, where researchers at the J. Craig Venter Institute synthesized an artificial bacterial genome back in 2008. From the Wiki page:
To identify the minimal genes required for life, each of the 482 genes of M. genitalium was individually deleted and the viability of the resulting mutants was tested. This resulted in the identification of a minimal set of 382 genes that theoretically should represent a minimal genome.[a 3] In 2008 the full set of M. genitalium genes was constructed in the laboratory with watermarks added to identify the genes as synthetic.
One of the newest works in this space was published just a few weeks ago in Jan 2025— part of the ‘Synthetic Yeast Genome Project’ and similarly hosted at the J. Craig Venter Institute —and is a degree more advanced, designing an existing yeast chromosome from scratch. The resulting chromosome, 902,994-bp long, reshuffles, redesigns, and removes existing genes.
As one may expect, genomic generations of this scale are painful to do in practice. Each chromosome was broken into chunks, then synthesized, tested, and debugged piece by piece. Genomic error correction had to be used to fix problems that arose after synthesis. The design phase itself took years of manual effort to decide which genes should stay, move, or be deleted. It should be no surprise that the Synthetic Yeast Genome Project has required over a decade of meticulous work by an international consortium of hundreds of scientist and still remains incomplete. While this 2025 paper wrapped up the final of 16 synthetic chromosomes to be designed, there still is work left to be done to actually integrate them into a single cell strain.
So…that’s the pitch for Evo 2. To do exactly this, but in a purely in-silico fashion and with a hopefully much higher hit-rate than other methods.
I still don’t get what the synthetic genome is doing that’s so different. Are they just adding in new, mostly innocuous nucleotides everywhere? Are there brand new biochemical processes being made? It feels like there’s a spectrum here between ‘new genomes that lead to functionally identical outcomes for the organism’ and ‘genuinely new life that could’ve never naturally evolved’.
I think it’s a mix.
Closer to the former side, the Synthetic Yeast Genome Project made it a point to drop transposons from the engineered yeast genome. Transposons are genetic elements that can move around and disrupt important coding or regulatory regions. In turn, they contribute genomic instability and make it harder to maintain a consistent genetic background for experiments or industrial applications. So removing them is genuinely novel and helpful for researchers! But you’re also not fundamentally changing the genome’s functionality. While transposons are present in nearly all life on Earth, they are more of an evolutionary byproduct that provides genetic variation over long timescales rather than something absolutely essential to the life of the host organism.
One far more interesting addition, closer to ‘never would’ve evolved naturally’ is the SCRaMbLE system (Synthetic Chromosome Rearrangement and Modification by LoxP-mediated Evolution) that was directly encoded into the synthetic yeast genome. This system introduces recombination sites throughout the synthetic yeast genome, allowing researchers to induce large-scale genome rearrangements on demand using Cre recombinase, effectively giving the yeast a built-in, programmable method of rapid evolution. SCRaMbLE enables controlled deletions, duplications, inversions, and rearrangements of genes in real-time, creating thousands of genome variants in a single experiment.
Of course, most created yeast strains will be completely dysfunctional or worse at doing [X] than the wildtype strain, but you only need to be lucky once! This just dramatically ramps up your ability to take shots on goal.
Did the Evo 2 authors actually do something like the aforementioned yeast genome redesign in the paper?
Well, no, the paper is very much focused on proof-of-concepts for now. What they did are three things:
One, valid mitochondria genome generation. Mitochondria have reasonably simple genomes—circular DNA molecules with a well-characterized set of protein-coding genes, tRNAs, and rRNAs, so they are a good first step if your goal is whole genome synthesis. Given a prompt of human mitchondrial DNA, Evo 2 infilled in the remaining, 16 kB worth of nucleotides. The generated genomes (of which they made 250) passed basic sanity checks, such as containing the expected number of genes and maintaining synteny (the physical location of the gene sequence) with natural mitochondrial genomes, like the below plot shows:
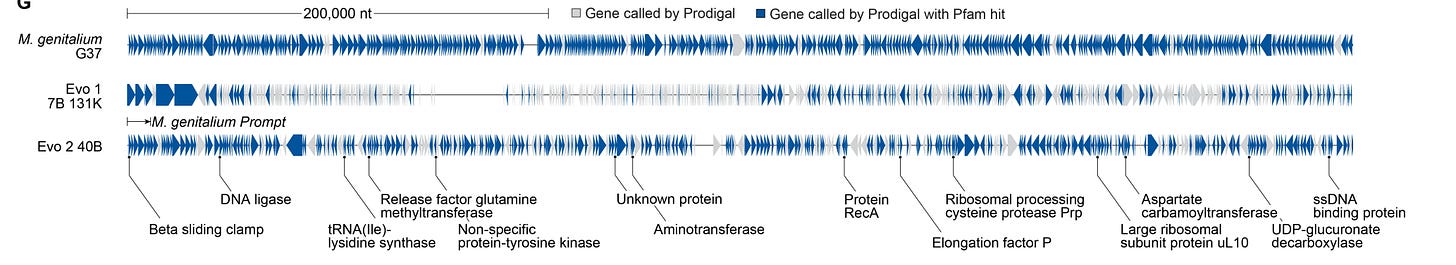
Two, prokaryotic genome generation. Specifically M. genitalium, the same organism redesigned back in 2008 that we previously discussed, and specifically known for its relatively small genome length (580 kB). Using the first 10.5 kb segment from the host genome, Evo 2 generated the remainder. The primary way that the resulting whole genome was assessed was by checking whether the predicted protein-coding regions contained Pfam ‘hits’, meaning that the proteins encoded by the Evo 2-generated genome matched known functional domains from real biological proteins. In turn, this serves as a proxy for biological plausibility.
In total, 70% of the Evo-2-generated genome had a Pfam hit. Predicted protein structures from the synthetic genome also had — according to ESMFold — structural analogs.
Three and finally, eukaryotic chromosome-scale genome generation. Specifically, Saccharomyces cerevisiae chromosome III, a 316 kb long chromosome from yeast. Using a 10.5 kb segment as a starting prompt, Evo 2 generated the remaining sequence, creating 20 fully synthetic yeast chromosomes. Just as with before, Pfam hits and structural assessments of genes with high Pfam hits were done, both of which looked good. That said, the authors note that it is unlikely that the generated yeast chromosome actually works, given that the density of tRNA (predicted via yeast-genome-specific tools unconnected to Evo 2) and absolute number of genes falls a fair bit below those of natural yeast genomes.
These feel like really strange ways to assess correctness of a generated genome. Protein domain matching? Structural analogs of predicted proteins from the genome? The genome doesn’t exist to produce proteins alone, it’s a huge, self-regulating conditional logic system! Pfam hits and structural analogs don’t capture any of that. They are, at best, heuristics for determining whether Evo 2 has generated something that looks like a genome, rather than one that actually functions as one. Why didn’t they just test this in a real cell?
Listen, we’re on the same page. But I think it’s worth considering how insanely complicated whole genome synthesis is.
Let’s say you wanted to personally test these DNA designs. You would’ve needed to assemble overlapping DNA fragments 10kB~ long, assemble them together inside of a cell, and then painstakingly screen for correct assemblies, because even with modern synthesis methods, errors creep in all the time. DNA synthesis isn’t perfect, and when you’re working with hundreds of thousands to millions of base pairs, the odds of introducing mutations or assembly failures are practically guaranteed. You’d then have to transfer the assembled genome into a viable host, which is an entire challenge on its own. You need to remove the original genome, prevent the cell from dying in the process, and hope that the synthetic genome actually starts functioning correctly.
It would’ve cost thousands of scientist hours and potentially millions of dollars. And what would’ve been the point? It’s almost certainly likely that the DNA generated by Evo 2 isn’t actually fully functional, I’m sure any of the authors would agree to that. But how nonfunctional? Is it entirely missing a whole set of extremely important transcription factors? Or, through sheer miracle, is it maybe just 10~ nucleotide mutations away from genuinely functioning? In either case, the only reaction from your cell would just be death, with you having learned no new information from the extremely expensive, multi-year endeavor.
I think one should look at the whole-DNA generation task that Evo 2 is doing as something that has like…no real functional way of being meaningfully evaluated beyond "does this look like somewhat like a genome?”. At least for now. You could get upset at this and the authors for not willing to really put skin in the game to test this out (for no reason, because they’d almost inevitably fail). But I think it’d be better to view this as an early poke into what really is a brand new scientific field, and brand new fields often lack easily accessible benchmarks for success.
Then what’s the point of genome generation? Why do any of this until we have those benchmarks?
I don’t control the levers of how R&D money is allocated, so my opinion is obviously meaningless. But I do think it is a very good idea for machine learning (ML) in biology to focus on problems where there is literally no alternative way to solve it. The current zeitgeist is people ignoring this advice and using ML to do CDR engineering with antibodies, which is questionable given that CDR’s are loopy, and you can assess protein loops at decently high-scales without needing any machine learning whatsoever (e.g. phage display). And what’s the end result of this? Maybe marginally better antibodies, that’s it. Nothing crazy happens as a result of someone really solving this.
On the other hand, if you can do reliable genome generation, you can create plants that sequester carbon at 1000x the typical rate, bacterial cells that can pump out antibodies fast enough to drive the cost of Humira down to pennies, and so on. But there is no other scalable way to do it at all without ML. If you wanted to rationally design a whole genome, you’d have to manually select every gene, every regulatory element, every replication origin, every transcription factor binding site, and then somehow figure out how all of them interact across a multi-megabase sequence. We’re clearly able to poke at very basic versions of this, but doing anything genuinely complicated is simply not feasible with human intuition alone, or at least requires so many decades to do as to not be scalable.
Now, fairly, it clearly seems to be infeasible for machines right now too, but at least ML is capable of trying—of exploring the combinatorial space of possible genomes in a way that humans fundamentally cannot. The alternative isn’t “do it manually”; it’s not doing it at all! And it’s clearly something worth doing.
So, in case you’re wondering if I’m a DNA language model supporter or hater, I’m officially counting myself in the former group, despite having starting these series starting at the latter. I just hope someone invents better ways to validate the results of them. This area just feels very much like something I’d want a blue sky, biology x computational research institution like Arc to focus on.
And that’s it! Thank you for reading, here is Part 1 again in case you’d like to see that again. And sorry for making this a 2-parter, the full thing would’ve been nearly 9,000 words long and that’s a bit much.
Is that it? There’s so much more covered in the paper! What about the whole mechanistic interpretability section?
I think it’s all very interesting, but it doesn’t feel worth talking about in depth, at least not yet. Not because it’s not cool, more because I cannot see the direct value of it. Similarly, while I find the mech interp stuff in the protein world cool, I’m still a bit unclear about the point. I’m sure I’ll be proven wrong about this very, very soon. Maybe that’ll be another essay in the future!






"if you can do reliable genome generation, you can create plants that sequester carbon at 1000x the typical rate" -- it seems that I'm still missing the point of these generative models even after reading your excellent essay as I don't understand how one could in principle request for a certain function from these models? All they know is generating natural-looking sequences and I'm failing to see how can we get from that to 1000x faster carbon sequestration?
One singular edit:
the genetic engineering you need to perform involves replacements much of a yeast cells genome -> the genetic engineering you need to perform involves replacements much of a yeast cell's genome