Announcements
Over the last two weeks, I released two articles:
Also, I’ll be hosting another NYC biotech meetup, this time on Wednesday, April 16th, at 7:30PM at Radegast Hall in Williamsburg. This will be the fourth meetup I’ve done in NYC, and there usually triple-digit attendees of extremely interesting people. You should come!
Finally, I had hoped a podcast would come out this month, but some delays have pushed it off. There should be one in April! If we’re lucky, maybe even two…
Links
Colossal Biosciences genetically edited a bunch of mice! Specifically to give them wholly-mammoth-esque phenotypes in their fur, which also made them very cute. Kenny from Latchbio had a great criticism of it (given below), but I still find this work cool; no huge biological advancement, but still strong technical execution on something hard + sets them up for more advanced work.
Corin Wagen wrote a wonderful post about the state of binding-affinity prediction using a Gödel, Escher, Bach-esque setup. It took me awhile to come back to this to read it, but it was surprisingly funny + informative, highly recommend!
All-atom diffusion transformers for periodic and non-periodic materials and molecules. Cool work! Currently only unconditional generation is available, but the sampled materials seem to meet in-silico benchmarks for validity quite well. This is an aside, but I’m not super familiar with material designs, so my mind immediately leaps to the controversy over the material design stuff that came from Deepmind in late 2023. What was the issue there? Kinda…nothing? At least no criticism that would be alien to someone familiar with how these types of models work. So likely little of it transfers to this new work.
Reticular dropped a web portal to do mechanistic interpretability on any given protein (using ESMFold). Here is a (AI-generated) thread over it.
Learning the language of protein-protein interactions. From the abstract: Here, we introduce MINT, a PLM specifically designed to model sets of interacting proteins in a contextual and scalable manner. Using unsupervised training on a large curated PPI dataset derived from the STRING database, MINT outperforms existing PLMs in diverse tasks relating to protein–protein interactions, including binding affinity prediction and estimation of mutational effects. Instinctively, I find the results here really surprising; I’ve always mentally modeled STRING as more of a protein-association thing. All it tells you is that a set of proteins are roughly found in the same place, not that they directly interact with each other. But, empirically, I suppose that if you have a large enough dataset of this sort of ‘probably interacting’ proteins, you can derive some very useful binding signal from it. Perhaps an example of unintentional generation 3 data?
NewLimit dropped their monthly update! Lots of good results on transcription factor discovery, but I think the most interesting bit is that their in-silico protein models: ‘These models take in a representation of an old cell state and a set of TFs and predict the effect of partial reprogramming on cell age and type….Our models can now explain more than half of the variation in cell age effects for unseen TF sets..’. I think it’s an open question how much models here can be pushed for transcription factor discovery (given how much of an issue OOD predictions are for protein models), looking forwards to an ML paper someday!
This isn’t scientifically interesting, but this animation by Profluent is really nice. There is an even better version on the homepage of their website.
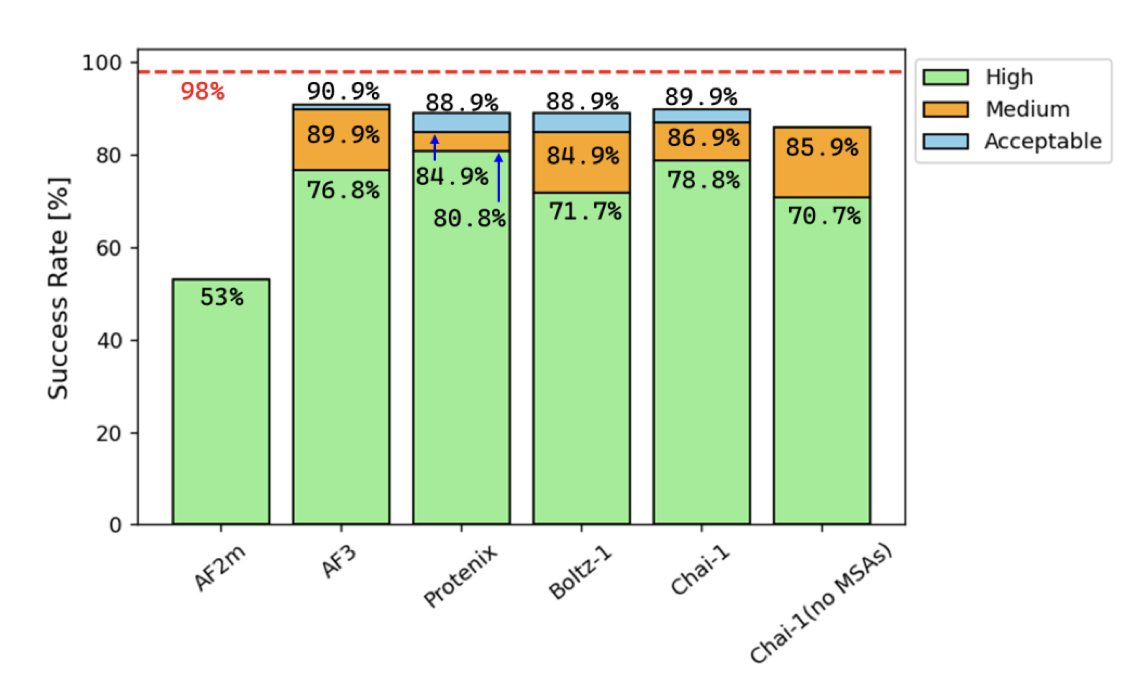
Benchmarking AlphaFold3-like Methods for Protein-Peptide Complex Prediction. Success rate here is given by 'ratio of the number of native interface contact residue pairs retained in the predicted model to the total number of native complex interface contact residues in the experimental crystal structure'. Crazy if true!
Jobs
Also feel free to reach out to her at lada@epilabs.co
Another longevity company, NewLimit, is aggressively hiring senior ML people in the Bay Area.
Asimov Press — a fantastic online journal — is hiring a part-time researcher to help write articles!




I am excited by MINT but I am not sure if they used association data. “We start with 2.4 billion protein-protein interactions (PPIs) comprising 59.3 million unique protein sequences classified as physical links from the STRING database.” I believe physical links are those where there is evidence of binding or a complex forming.
(1) thanks for the shout-out!
(2) I'm not aware of super big controversy re: GNoME, it seems like a solid piece of work. the comment you linked seems about right. most of the complaints are about presentation, framing, and so forth (as opposed to the previous self-driving materials exploration work, where many of the XRD data were just incompetently analyzed and flat-out wrong).
(3) re: AF3 protein–peptide modeling, at least for small molecule ligands it seems that OOD performance is not great (cf. the papers I cite in my Achilles/tortoise post). I'm curious about if the observed good performance is a function of data leakage, or if cofolding methods are just better at peptides. it's very plausible that these methods are better at handling peptides, since there's much more data (and much less diversity), and one would hope that some of the classic AlphaFold ML performance would translate!